18 KiB
title, keywords, last_updated, summary, sidebar, permalink
| title | keywords | last_updated | summary | sidebar | permalink |
|---|---|---|---|---|---|
| Archive & PSI Data Catalog | linux, archive, data catalog, archiving, lts, tape, long term storage, ingestion, datacatalog | 31 January 2020 | This document describes how to use the PSI Data Catalog for archiving Merlin7 data. | merlin7_sidebar | /merlin7/archive.html |
PSI Data Catalog as a PSI Central Service
PSI provides access to the Data Catalog for long-term data storage and retrieval. Data is stored on the PetaByte Archive at the Swiss National Supercomputing Centre (CSCS).
The Data Catalog and Archive is suitable for:
- Raw data generated by PSI instruments
- Derived data produced by processing some inputs
- Data required to reproduce PSI research and publications
The Data Catalog is part of PSI's effort to conform to the FAIR principles for data management. In accordance with this policy, data will be publicly released under CC-BY-SA 4.0 after an embargo period expires.
The Merlin cluster is connected to the Data Catalog. Hence, users archive data stored in the
Merlin storage under the /data directories (currentlyi, /data/user and /data/project).
Archiving from other directories is also possible, however the process is much slower as data
can not be directly retrieved by the PSI archive central servers (central mode), and needs to
be indirectly copied to these (decentral mode).
Archiving can be done from any node accessible by the users (usually from the login nodes).
{{site.data.alerts.tip}} Archiving can be done in two different ways:
'Central mode': Possible for the user and project data directories, is the
fastest way as it does not require remote copy (data is directly retreived by central AIT servers from Merlin
through 'merlin-archive.psi.ch').
'Decentral mode': Possible for any directory, is the slowest way of archiving as it requires
to copy ('rsync') the data from Merlin to the central AIT servers.
{{site.data.alerts.end}}
Procedure
Overview
Below are the main steps for using the Data Catalog.
- Ingest the dataset into the Data Catalog. This makes the data known to the Data Catalog system at PSI:
- Prepare a metadata file describing the dataset
- Run
datasetIngestorscript - If necessary, the script will copy the data to the PSI archive servers
- Usually this is necessary when archiving from directories other than
/data/useror/data/project. It would be also necessary when the Merlin export server (merlin-archive.psi.ch) is down for any reason.
- Usually this is necessary when archiving from directories other than
- Archive the dataset:
- Visit https://discovery.psi.ch
- Click
Archivefor the dataset - The system will now copy the data to the PetaByte Archive at CSCS
- Retrieve data from the catalog:
- Find the dataset on https://discovery.psi.ch and click
Retrieve - Wait for the data to be copied to the PSI retrieval system
- Run
datasetRetrieverscript
- Find the dataset on https://discovery.psi.ch and click
Since large data sets may take a lot of time to transfer, some steps are designed to happen in the background. The discovery website can be used to track the progress of each step.
Account Registration
Two types of account permit access to the Data Catalog. If your data was collected at a beamline, you may
have been assigned a p-group (e.g. p12345) for the experiment. Other users are assigned a-group
(e.g. a-12345).
Groups are usually assigned to a PI, and then individual user accounts are added to the group. This must be done under user request through PSI Service Now. For existing a-groups and p-groups, you can follow the standard central procedures. Alternatively, if you do not know how to do that, follow the Merlin7 Requesting extra Unix groups procedure, or open a PSI Service Now ticket.
Documentation
Accessing the Data Catalog is done through the SciCat software. Documentation is here: ingestManual.
Loading datacatalog tools
The latest datacatalog software is maintained in the PSI module system. To access it from the Merlin systems, run the following command:
module load datacatalog
It can be done from any host in the Merlin cluster accessible by users. Usually, login nodes will be the nodes used for archiving.
Finding your token
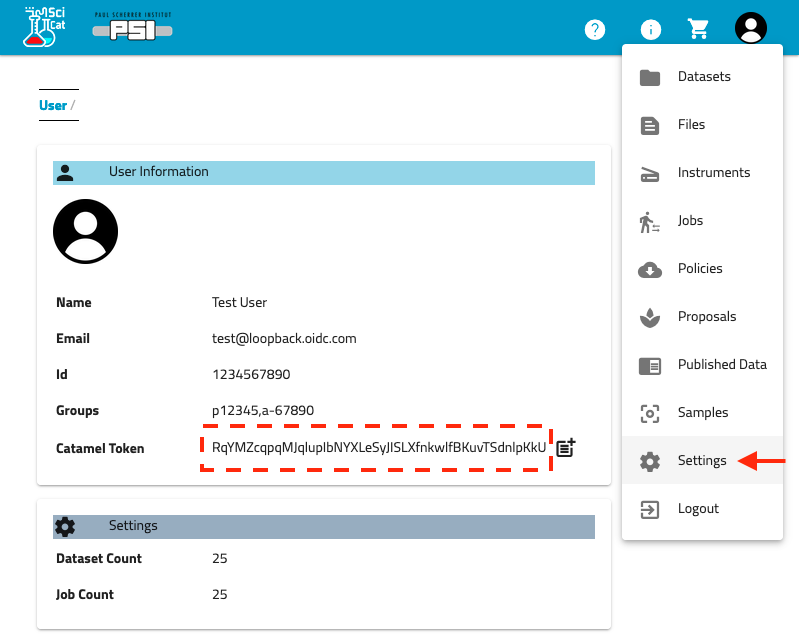
As of 2022-04-14 a secure token is required to interact with the data catalog. This is a long random string that replaces the previous user/password authentication (allowing access for non-PSI use cases). This string should be treated like a password and not shared.
- Go to discovery.psi.ch
- Click 'Sign in' in the top right corner. Click the 'Login with PSI account' and log in on the PSI login1. page.
- You should be redirected to your user settings and see a 'User Information' section. If not, click on1. your username in the top right and choose 'Settings' from the menu.
- Look for the field 'Catamel Token'. This should be a 64-character string. Click the icon to copy the1. token.
You will need to save this token for later steps. To avoid including it in all the commands, I suggest saving it to an environmental variable (Linux):
$ SCICAT_TOKEN=RqYMZcqpqMJqluplbNYXLeSyJISLXfnkwlfBKuvTSdnlpKkU
(Hint: prefix this line with a space to avoid saving the token to your bash history.)
Tokens expire after 2 weeks and will need to be fetched from the website again.
Ingestion
The first step to ingesting your data into the catalog is to prepare a file describing what data you have. This is called
metadata.json, and can be created with a text editor (e.g. vim). It can in principle be saved anywhere,
but keeping it with your archived data is recommended. For more information about the format, see the 'Bio metadata'
section below. An example follows:
{
"principalInvestigator": "albrecht.gessler@psi.ch",
"creationLocation": "/PSI/EMF/JEOL2200FS",
"dataFormat": "TIFF+LZW Image Stack",
"sourceFolder": "/gpfs/group/LBR/pXXX/myimages",
"owner": "Wilhelm Tell",
"ownerEmail": "wilhelm.tell@psi.ch",
"type": "raw",
"description": "EM micrographs of amygdalin",
"ownerGroup": "a-12345",
"scientificMetadata": {
"description": "EM micrographs of amygdalin",
"sample": {
"name": "Amygdalin beta-glucosidase 1",
"uniprot": "P29259",
"species": "Apple"
},
"dataCollection": {
"date": "2018-08-01"
},
"microscopeParameters": {
"pixel size": {
"v": 0.885,
"u": "A"
},
"voltage": {
"v": 200,
"u": "kV"
},
"dosePerFrame": {
"v": 1.277,
"u": "e/A2"
}
}
}
}
It is recommended to use the ScicatEditor for creating metadata files. This is a browser-based tool specifically for ingesting PSI data. Using the tool avoids syntax errors and provides templates for common data sets and options. The finished JSON file can then be downloaded to merlin or copied into a text editor.
Another option is to use the SciCat graphical interface from NoMachine. This provides a graphical interface for selecting data to archive. This is particularly useful for data associated with a DUO experiment and p-group. Type SciCat`` to get started after loading the datacatalog`` module. The GUI also replaces the the command-line ingestion described below.
The following steps can be run from wherever you saved your metadata.json. First, perform a "dry-run" which will check the metadata for errors:
datasetIngestor --token $SCICAT_TOKEN metadata.json
It will ask for your PSI credentials and then print some info about the data to be ingested. If there are no errors, proceed to the real ingestion:
datasetIngestor --token $SCICAT_TOKEN --ingest --autoarchive metadata.json
You will be asked whether you want to copy the data to the central system:
- If you are on the Merlin cluster and you are archiving data from
/data/useror/data/project, answer 'no' since the data catalog can directly read the data. - If you are on a directory other than
/data/userand ``/data/project, or you are on a desktop computer, answer 'yes'. Copying large datasets to the PSI archive system may take quite a while (minutes to hours).
If there are no errors, your data has been accepted into the data catalog! From now on, no changes should be made to the ingested data. This is important, since the next step is for the system to copy all the data to the CSCS Petabyte archive. Writing to tape is slow, so this process may take several days, and it will fail if any modifications are detected.
If using the --autoarchive option as suggested above, your dataset should now be in the queue. Check the data catalog:
https://discovery.psi.ch. Your job should have status 'WorkInProgress'. You will receive an email when the ingestion
is complete.
If you didn't use --autoarchive, you need to manually move the dataset into the archive queue. From discovery.psi.ch, navigate to the 'Archive'
tab. You should see the newly ingested dataset. Check the dataset and click Archive. You should see the status change from datasetCreated to
scheduleArchiveJob. This indicates that the data is in the process of being transferred to CSCS.
After a few days the dataset's status will change to datasetOnAchive indicating the data is stored. At this point it is safe to delete the data.
Useful commands
Running the datasetIngestor in dry mode (without --ingest) finds most errors. However, it is sometimes convenient to find potential errors
yourself with simple unix commands.
Find problematic filenames
find . -iregex '.*/[^/]*[^a-zA-Z0-9_ ./-][^/]*'=
Find broken links
find -L . -type l
Find outside links
find . -type l -exec bash -c 'realpath --relative-base "`pwd`" "$0" 2>/dev/null |egrep "^[./]" |sed "s|^|$0 ->|" ' '{}' ';'
Delete certain files (use with caution)
# Empty directories
find . -type d -empty -delete
# Backup files
find . -name '*~' -delete
find . -name '*#autosave#' -delete
Troubleshooting & Known Bugs
-
The following message can be safely ignored:
key_cert_check_authority: invalid certificate Certificate invalid: name is not a listed principalIt indicates that no kerberos token was provided for authentication. You can avoid the warning by first running kinit (PSI linux systems).
-
For decentral ingestion cases, the copy step is indicated by a message
Running [/usr/bin/rsync -e ssh -avxz .... It is expected that this step will take a long time and may appear to have hung. You can check what files have been successfully transfered using rsync:rsync --list-only user_n@pb-archive.psi.ch:archive/UID/PATH/where UID is the dataset ID (12345678-1234-1234-1234-123456789012) and PATH is the absolute path to your data. Note that rsync creates directories first and that the transfer order is not alphabetical in some cases, but it should be possible to see whether any data has transferred.
-
There is currently a limit on the number of files per dataset (technically, the limit is from the total length of all file paths). It is recommended to break up datasets into 300'000 files or less.
-
If it is not possible or desirable to split data between multiple datasets, an alternate work-around is to package files into a tarball. For datasets which are already compressed, omit the -z option for a considerable speedup:
tar -f [output].tar [srcdir]Uncompressed data can be compressed on the cluster using the following command:
sbatch /data/software/Slurm/Utilities/Parallel_TarGz.batch -s [srcdir] -t [output].tar -nRun /data/software/Slurm/Utilities/Parallel_TarGz.batch -h for more details and options.
-
Sample ingestion output (datasetIngestor 1.1.11)
[Show Example]: Sample ingestion output (datasetIngestor 1.1.11)
/data/project/bio/myproject/archive $ datasetIngestor -copy -autoarchive -allowexistingsource -ingest metadata.json
2019/11/06 11:04:43 Latest version: 1.1.11
2019/11/06 11:04:43 Your version of this program is up-to-date
2019/11/06 11:04:43 You are about to add a dataset to the === production === data catalog environment...
2019/11/06 11:04:43 Your username:
user_n
2019/11/06 11:04:48 Your password:
2019/11/06 11:04:52 User authenticated: XXX
2019/11/06 11:04:52 User is member in following a or p groups: XXX
2019/11/06 11:04:52 OwnerGroup information a-XXX verified successfully.
2019/11/06 11:04:52 contactEmail field added: XXX
2019/11/06 11:04:52 Scanning files in dataset /data/project/bio/myproject/archive
2019/11/06 11:04:52 No explicit filelistingPath defined - full folder /data/project/bio/myproject/archive is used.
2019/11/06 11:04:52 Source Folder: /data/project/bio/myproject/archive at /data/project/bio/myproject/archive
2019/11/06 11:04:57 The dataset contains 100000 files with a total size of 50000000000 bytes.
2019/11/06 11:04:57 creationTime field added: 2019-07-29 18:47:08 +0200 CEST
2019/11/06 11:04:57 endTime field added: 2019-11-06 10:52:17.256033 +0100 CET
2019/11/06 11:04:57 license field added: CC BY-SA 4.0
2019/11/06 11:04:57 isPublished field added: false
2019/11/06 11:04:57 classification field added: IN=medium,AV=low,CO=low
2019/11/06 11:04:57 Updated metadata object:
{
"accessGroups": [
"XXX"
],
"classification": "IN=medium,AV=low,CO=low",
"contactEmail": "XXX",
"creationLocation": "XXX",
"creationTime": "2019-07-29T18:47:08+02:00",
"dataFormat": "XXX",
"description": "XXX",
"endTime": "2019-11-06T10:52:17.256033+01:00",
"isPublished": false,
"license": "CC BY-SA 4.0",
"owner": "XXX",
"ownerEmail": "XXX",
"ownerGroup": "a-XXX",
"principalInvestigator": "XXX",
"scientificMetadata": {
...
},
"sourceFolder": "/data/project/bio/myproject/archive",
"type": "raw"
}
2019/11/06 11:04:57 Running [/usr/bin/ssh -l user_n pb-archive.psi.ch test -d /data/project/bio/myproject/archive].
key_cert_check_authority: invalid certificate
Certificate invalid: name is not a listed principal
user_n@pb-archive.psi.ch's password:
2019/11/06 11:05:04 The source folder /data/project/bio/myproject/archive is not centrally available (decentral use case).
The data must first be copied to a rsync cache server.
2019/11/06 11:05:04 Do you want to continue (Y/n)?
Y
2019/11/06 11:05:09 Created dataset with id 12.345.67890/12345678-1234-1234-1234-123456789012
2019/11/06 11:05:09 The dataset contains 108057 files.
2019/11/06 11:05:10 Created file block 0 from file 0 to 1000 with total size of 413229990 bytes
2019/11/06 11:05:10 Created file block 1 from file 1000 to 2000 with total size of 416024000 bytes
2019/11/06 11:05:10 Created file block 2 from file 2000 to 3000 with total size of 416024000 bytes
2019/11/06 11:05:10 Created file block 3 from file 3000 to 4000 with total size of 416024000 bytes
...
2019/11/06 11:05:26 Created file block 105 from file 105000 to 106000 with total size of 416024000 bytes
2019/11/06 11:05:27 Created file block 106 from file 106000 to 107000 with total size of 416024000 bytes
2019/11/06 11:05:27 Created file block 107 from file 107000 to 108000 with total size of 850195143 bytes
2019/11/06 11:05:27 Created file block 108 from file 108000 to 108057 with total size of 151904903 bytes
2019/11/06 11:05:27 short dataset id: 0a9fe316-c9e7-4cc5-8856-e1346dd31e31
2019/11/06 11:05:27 Running [/usr/bin/rsync -e ssh -avxz /data/project/bio/myproject/archive/ user_n@pb-archive.psi.ch:archive
/0a9fe316-c9e7-4cc5-8856-e1346dd31e31/data/project/bio/myproject/archive].
key_cert_check_authority: invalid certificate
Certificate invalid: name is not a listed principal
user_n@pb-archive.psi.ch's password:
Permission denied, please try again.
user_n@pb-archive.psi.ch's password:
/usr/libexec/test_acl.sh: line 30: /tmp/tmpacl.txt: Permission denied
/usr/libexec/test_acl.sh: line 30: /tmp/tmpacl.txt: Permission denied
/usr/libexec/test_acl.sh: line 30: /tmp/tmpacl.txt: Permission denied
/usr/libexec/test_acl.sh: line 30: /tmp/tmpacl.txt: Permission denied
/usr/libexec/test_acl.sh: line 30: /tmp/tmpacl.txt: Permission denied
...
2019/11/06 12:05:08 Successfully updated {"pid":"12.345.67890/12345678-1234-1234-1234-123456789012",...}
2019/11/06 12:05:08 Submitting Archive Job for the ingested datasets.
2019/11/06 12:05:08 Job response Status: okay
2019/11/06 12:05:08 A confirmation email will be sent to XXX
12.345.67890/12345678-1234-1234-1234-123456789012
Publishing
After datasets are are ingested they can be assigned a public DOI. This can be included in publications and will make the datasets on http://doi.psi.ch.
For instructions on this, please read the 'Publish' section in the ingest manual.
Retrieving data
Retrieving data from the archive is also initiated through the Data Catalog. Please read the 'Retrieve' section in the ingest manual.
Further Information
- PSI Data Catalog
- Full Documentation
- Published Datasets (doi.psi.ch)
- Data Catalog PSI page
- Data catalog SciCat Software
- FAIR definition and SNF Research Policy
- Petabyte Archive at CSCS